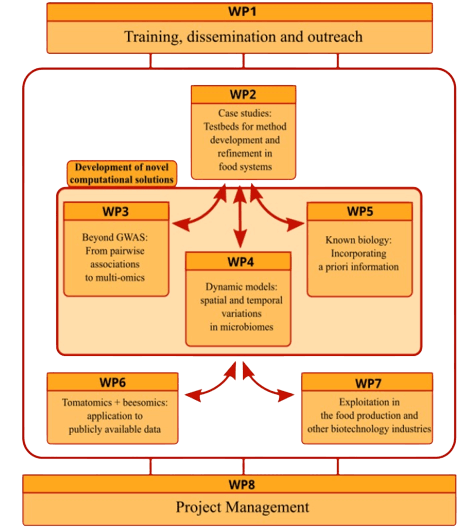
FindingPheno consists of eight work packages (WP) including:
- one management WP
- two structural WPs, and
- five technical WPs
Over 48 months the consortium will develop and implement novel technical and statistical methods, and provide access to easily accessible software packages using statistical methods, as well as undertake outreach and wide dissemination of project results.